Problem statement
Recent developments in neural tracing have made it possible to stain the entire axonal arborization of individual long-range projecting neurons. While advances in whole-brain scanning are being made, most labs rely on stained tissue sections in which axonal fragments are manually traced using Neurolucida (MBF Bioscience) or similar software. Fragments in subsequent sections need to be stitched: the sections need to be aligned and matching fragments must be connected. This tedious task consumes more time than the tracing itself.
Stitching pipeline
- A variant of the Dercksen et al. (2009, doi 10.1109/ISBI.2009.5193216) algorithm is used that aligns high endings of fragments to low endings of fragments in the adjacent section.
- This resulting set of putative links are loaded into a visual stitching tool, in which about 5 to 10 putative links per set of adjacent sections are confirmed to become a 'stitch'.
- With the confirmed links as constraints, step 1 is repeated, resulting in a final set of putative links that can be confirmed as a group.
- In the visual stitching tool, unpaired fragments are confirmed as 'terminals', or the qualification 'low ending' or 'high ending' is corrected.
Implementation
Steps 1 and 3 are implemented in Python and C, and put together in a Jupyter Notebook. Steps 2 and 4 use the web-based stitching tool.
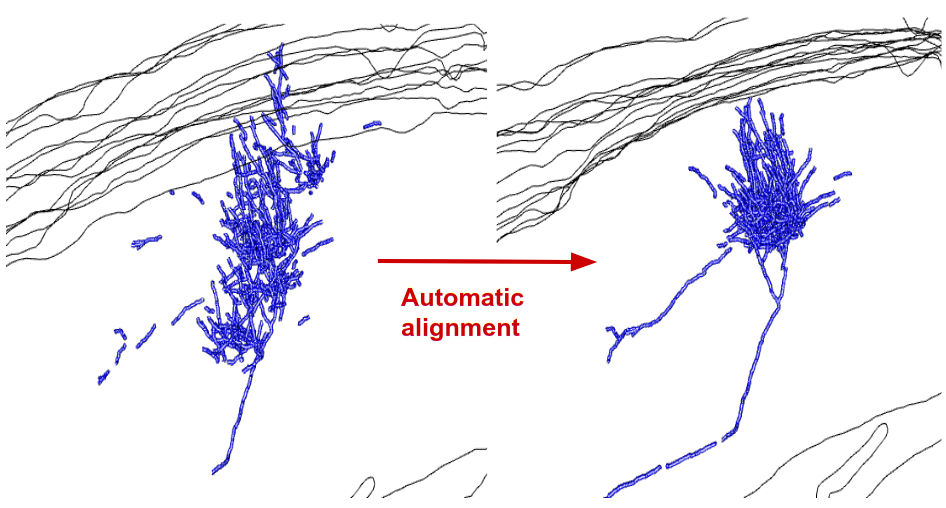 *Figure: Axonal arborization before and after applying the modified Dercksen algorithm. Especially in areas with many fragments the result is very good. If only a few fragments are available in adjacent sections, an alternative type of alignment is needed, for instance based on tissue contours or the actual tissue.
*Figure: Axonal arborization before and after applying the modified Dercksen algorithm. Especially in areas with many fragments the result is very good. If only a few fragments are available in adjacent sections, an alternative type of alignment is needed, for instance based on tissue contours or the actual tissue.